Diabetes Data Processing
In the NGI funded MyPCH project OwnYourData developed several technologies for secure and traceable data exchange of diabetes data: Digital Watermarking, Semantic Annotation, and Data Traceability. In addition, we also participated in a MyData Health initiative to write a feature article for the European Medical Writers Association (EMWA) and we are proud to announce that the article written by 14 individuals from 9 countries around the 6 MyData principles is already published and public available via the EMWA journal website – see section “Data Interoperability” and “Establishing trust between stakeholders for health data use” for example use cases of Semantic Containers.
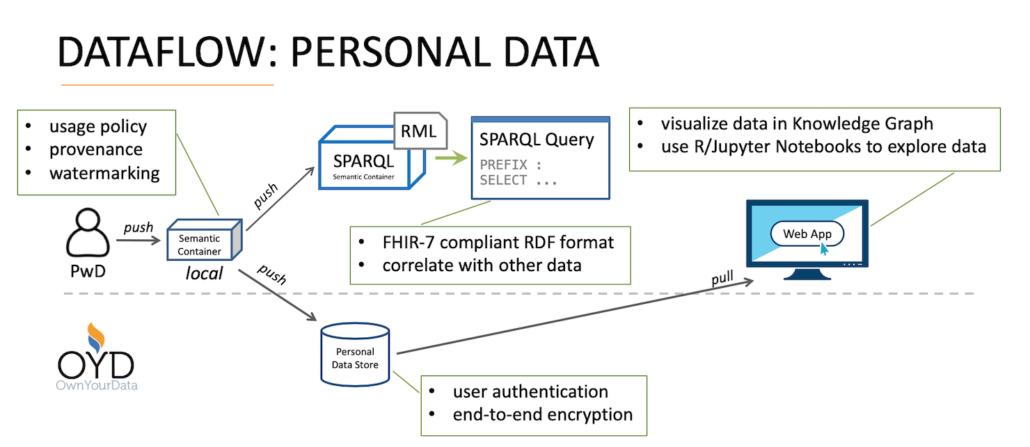
In this blog post we cover the successful integration of diabetes data into the OwnYourData Data Vault. In this data flow, persons with diabetes (Pwd) can not only transfer their data to a Personal Data Store but also perform SPARQL queries to combine their diabetes data with public information – more information and examples are available here.
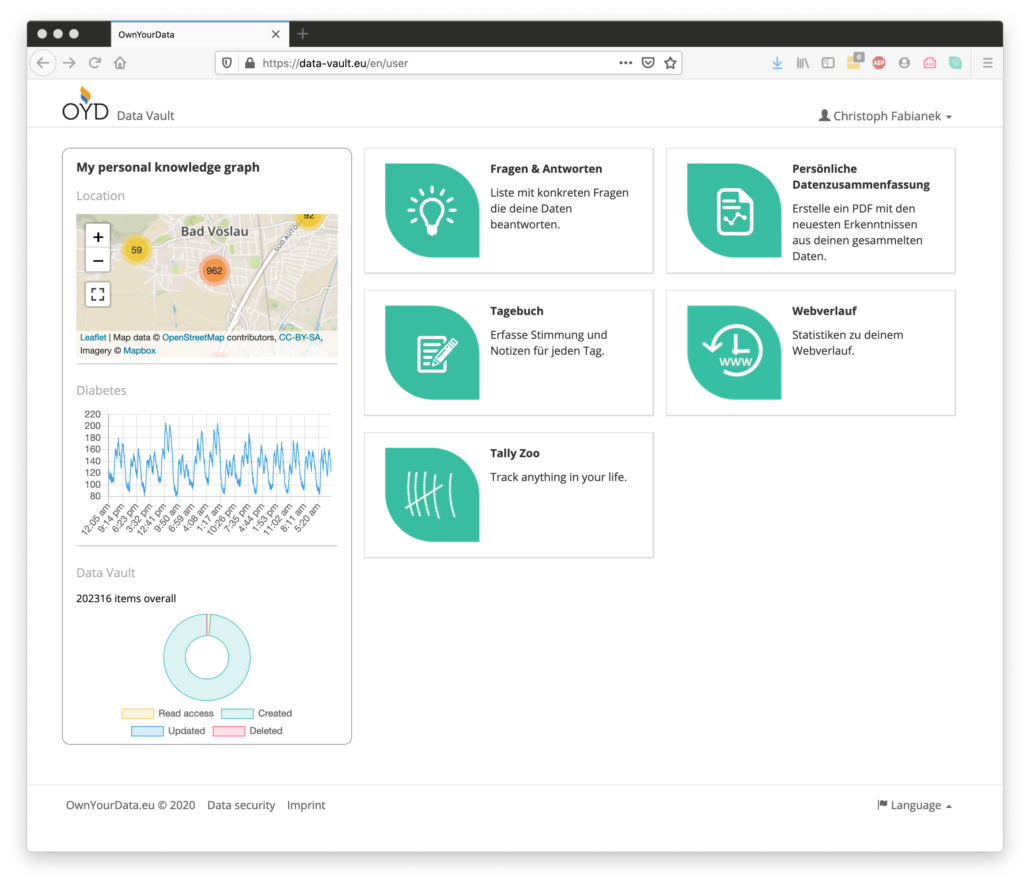
A special feature in the OwnYourData Data Vault is the Personal Knowledge Graph shown on the left side of the main screen. It compiles available data from the respective user and presents the information in a clear form. The screenshot below shows information about recent GPS Data, blood sugar levels over the course of a few days, as well as record numbers overall.
Beyond information in the Personal Knowledge Graph plugins allow further data exploration. In the course of the MyPCH project however, we decided to use existing tools like R or Jupyter Notebooks to provide more sophisticated visualization and analysis mechanisms. The R-Notebook available on Github is an example how to retrieve and decrypt information in the Data Vault and compile a report.
If you have any questions using Diabetes data with Semantic Containers or within the OwnYourData Data Vault don’t hesitate to contact us as support@ownyourdata.eu.